Suneel Apte Laboratory
-
Suneel Apte Laboratory
- Principal Investigator
- Research
- Our Team
- Publications
- Patents
- Research News
Research
The Apte laboratory works on proteases, enzymes which break down proteins and thus have a crucial function in both health and disease. We are specifically interested in the activity of proteases (which is termed proteolysis) against extracellular matrix, which is the material that holds cells together, constitutes their primary microenvironment, defines their polarity and regulates their behavior.
Extracellular matrix proteolysis is essential for normal organ formation and function and occurs continuously within the body. However, both too much or too little of it can impair organ function in various ways. After defining the roles of many newly discovered proteases in mammalian embryonic development, we are now interested in proteolysis more broadly and in some of the most significant health challenges facing patients. We investigate proteolysis in different forms of arthritis (e.g., osteoarthritis, inflammatory arthritis) and cardiovascular disorders such as heart failure and aortic aneurysms. Our objective is to identify new disease pathways, biomarkers and drugs and develop a comprehensive framework for understanding proteolysis across organs and disease ecosystems.
Biography
Suneel Apte, MBBS, D Phil, earned his medical degree from the University of Bombay (now the University of Mumbai), India. He interrupted his clinical training as an orthopedic surgeon to pursue a D. Phil degree at the University of Oxford, where he was also a Rhodes Scholar, and was mentored by John Kenwright, BM, MCh, MA, FRCS. He undertook post-doctoral training with Bjorn R. Olsen, MD, PhD, at Harvard Medical School before establishing an independent research program at the Cleveland Clinic, where he now has an appointment as Staff.
Education & Professional Highlights
|
University of Bombay, India |
M.B.B.S. |
1985 |
Clinical Medicine |
|
University of Oxford, United Kingdom |
D.Phil. |
1990 |
Clinical Medicine |
|
Harvard Medical School, Boston, MA |
Post-doctoral training |
1994 |
Molecular Biology |
Appointments
1996 Assistant Staff - Department of Biomedical Engineering & Department of Orthopaedic Surgery
2001 Associate Staff - Department of Biomedical Engineering & Department of Orthopaedic Surgery
2006 Staff - Department of Biomedical Engineering & Department of Orthopaedic Surgery
Prior Appointments
1994-1996 Instructor, Dept. of Cell Biology, Harvard Medical School, Boston, MA
1987-1989 Honorary Registrar, Nuffield Department of Orthopaedic Surgery,
Nuffield Orthopaedic Center, Oxford, UK
1984-1986 Intern and Resident in Orthopaedics, King Edward VII Memorial Hospital,
Bombay, India
Professional Highlights
Dr. Apte is the Past-President of the International Society for Matrix Biology, a Past-President of the American Society for Matrix Biology, whose biennial conference he organized in 2014 and was Chair of the 2013 Gordon Research Conference on Metalloproteinases. He served two terms as Editor with the Journal of Biological Chemistry, and is currently an Editor of Matrix Biology, and the Journal's companion title, Matrix Biology Plus. He is a former American Heart Association-Paul G. Allen Frontiers Group Distinguished Investigator. He has been a reviewer on several NIH, NASA and Arthritis Foundation study sections, a scientific advisory board member for the The Marfan Foundation, Shriners Hospitals for Children, and The Kennedy Institute for Rheumatology and a member of the Board of Directors of the Orthopedic Research Society. He chaired the International Scientific Advisory Board for a BBSRC sLoLa award made jointly to the Universities of Manchester and Bristol, UK. The Apte laboratory has trained numerous post-doctoral fellows, medical and doctoral students, undergraduates and technicians who have gone on to careers as researchers in the academic or industry sectors, medicine and allied professions, science publishing and patent law.
Research
Overview
The Apte Lab conducts fundamental research on extracellular matrix and the proteases that remodel it. Extracellular matrix is very important because it provides a structural scaffold for every tissue and organ and regulates cell behavior.
The laboratory's work spans organ, tissue, cell and molecular scales to determine how extracellular matrix turnover contributes to human diseases. The matrix is continuously remodeled by balanced production and breakdown (proteolysis), the latter undertaken by matrix degrading proteases. The ADAMTS proteases, a focus of our laboratory, are important participants in this process. In addition to uncovering their roles in human disease, we also broadly investigate extracellular matrix proteolysis in disorders such as osteoarthritis, inflammatory arthritis, aortic aneurysms and heart failure, which are responsible for a large proportion of the disease burden in any population.
Some of our recent research publications are highlighted below. Please follow the links to access the full articles.
Research Support
The laboratory's current research is supported by the National Institutes of Health (NIH).
The lab gratefully acknowledges support received over the years from several NIH institutes, including NIAMS, NEI, NICHD and the NIH-NHLBI Program of Excellence in Glycosciences, the Allen Distinguished Investigator Program, through support made possible by The Paul G. Allen Frontiers Group and the American Heart Association, as well as The Marfan Foundation, The Arthritis Foundation, Sabrina's Foundation, the Ines Mandl Research Foundation, Cleveland Clinic Velosano and the Cleveland Clinic Catalyst Award.
Database Creation
DICED (Database of Identified Cleavage Sites Endemic to Disease States) is a web interface that offers a user-friendly portal to N-terminomics data. This was created in collaboration by Jayadev Joshi, Sumit Bhutada, Daniel Martin, Joyce Guzowski, Daniel Blankenberg and Suneel Apte at the Cleveland Clinic.
Our Team
Selected Publications
View publications for Suneel Apte, MBBS, DPhil
(Disclaimer: This search is powered by PubMed, a service of the U.S. National Library of Medicine. PubMed is a third-party website with no affiliation with Cleveland Clinic.)
Names of the Apte lab members are written in bold text
Publications in 2025
(Book Chapter). Traboulsi EI, Ghali, N, Apte, SS. Ectopia lentis and associated systemic disease. In, Traboulsi EI (Ed), Genetic Diseases of the Eye, 3rd Edition, Oxford university Press., New York, pp 849-874, ISBN 9780197659410.
Taye N, Karoulias SZ, Balic Z, Wang LW, Willard BB, Martin D, Richard D, Okamoto AS, Capellini TD, Apte SS, Hubmacher D. Combined ADAMTS10 and ADAMTS17 inactivation exacerbates bone shortening and skin phenotypes. Life Sci Alliance. 2025 Oct 1;8(12):e202503232. doi: 10.26508/lsa.202503232. Print 2025 Dec.PMID: 41034152
Martin DR, Sardelli G, Burkhard T, Fowkes MM, Minns AF, Roberta Moschini R, Del Corso A, de Groot R, Apte SS*, Santamaria S*. Characterization of ADAMTS9 proteoglycanase activity: comparison with ADAMTS1, ADAMTS4 and ADAMTS5. J Biol Chem, DOI: 10.1016/j.jbc.2025.110301
Joshi J, Bhutada S, Martin DR, Guzowski J, Blankenberg D, Apte SS. DICED (Database of Identified Cleavage Sites Endemic to Diseases States): A Searchable Web Interface for Terminomics/Degradomics. Proteomics. 2025 May 12:e202500007. doi: 10.1002/pmic.202500007
Westöö C, Mutgan AC, van der Have O, Mead TJ, Ahmed S, Lampei E, Koch CD, Norvik C, Aspberg A, Bech M, Peruzzi N, Brunnström H, Kwapiszewska G, Rådegran G, Apte SS, Tran-Lundmark K.Localization, Proteolytic Processing, and Binding Partners of Versican Isoforms in Vascular Lesions of Pulmonary Arterial Hypertension. J Histochem Cytochem. 2025 Mar-Apr;73(3-4):129-145. doi: 10.1369/00221554251331271
Mead T.J.,, Bhutada, S., Peruzzi N., Adegboye J., Seifert D.E., Cahill E., Drinko J., Donnellan E., Guggiliam A., Popovic Z., Griffin B., Tran-Lundmark K., Apte, S. S. ADAMTS7, a target in atherosclerosis, cooperates with its homolog ADAMTS12 to protect against myxomatous valve degeneration. Journal of Molecular and Cellular Cardiology Plus. 2025 Feb 22;11:100288. doi: 10.1016/j.jmccpl.2025.100288
(Review) Khan ST, Huffman N, Li X, Sharma A, Winalski CS, Ricchetti ET, Derwin K, Apte SS, Rotroff D, Saab C, Piuzzi NS. Pain Assessment in Osteoarthritis: Present Practices and Future Prospects Including the Use of Biomarkers and Wearable Technologies, and AI-Driven Personalized Medicine. J Orthop Res. 2025 Apr 9. doi: 10.1002/jor.26082
Publications in 2024
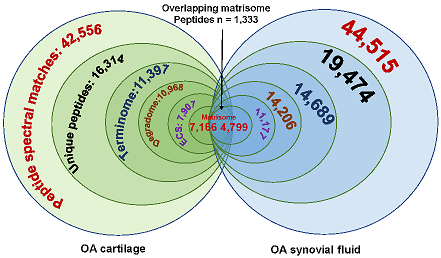
Bhutada S, Hoyle A, Piuzzi NS, Apte SS. Degradomics defines proteolysis information flow from human knee osteoarthritis cartilage to matched synovial fluid and the contributions of secreted proteases ADAMTS5, MMP13 and CMA1 to articular cartilage breakdown. Osteoarthritis Cartilage. 2024 Sep 16:S1063-4584(24)01397-9. doi: 10.1016/j.joca.2024.09.002. PMID: 39293776.
Kuczynski-Noyau L, Karmann S, Alberton P, Martinez-Corral I, Nampoothiri S, Sauvé F, Lhomme T, Quarta C, Apte SS, Bouret S, Aszodi A, Rasika S, Ciofi P, Dam J, Prévot V, Mattot V. A plastic aggrecan barrier modulated by peripheral energy state gates metabolic signal access to arcuate neurons. Nat Commun. 2024 Aug 7;15(1):6701. doi: 10.1038/s41467-024-50798-9. PMID: 39112471; PMCID: PMC11306556.
Mead TJ, Bhutada S, Foulcer SJ, Peruzzi N, Nelson CM, Seifert DE, Larkin J, Tran-Lundmark K, Filmus J, Apte SS. Combined genetic-pharmacologic inactivation of tightly linked ADAMTS proteases in temporally specific windows uncovers distinct roles for versican proteolysis and glypican-6 in cardiac development. Matrix Biology 2024 May 13: S0945-053X(24)00064-7. doi: https://doi.org/10.1016/j.matbio.2024.05.003.
Rypdal KB, Apte SS, Lunde IG. Emerging roles for the ADAMTS-like family of matricellular proteins in cardiovascular disease through regulation of the extracellular microenvironment. Mol Biol Rep. 2024 Feb 7;51(1):280. doi: 10.1007/s11033-024-09255-5. PMID: 38324186; PMCID: PMC10850197.
de Groot R, Folgado PB, Yamamoto K, Martin DR, Koch CD, Debruin D, Blagg S, Minns AF, Bhutada S, Ahnström J, Larkin J, Aspberg A, Önnerfjord P, Apte SS, Santamaria S. Cleavage of Cartilage Oligomeric Matrix Protein (COMP) by ADAMTS4 generates a neoepitope associated with osteoarthritis and other forms of degenerative joint disease. Matrix Biology 2024 Dec 11:S0945-053X(24)00147-1. doi: 10.1016/j.matbio.2024.12.005. Online ahead of print.PMID: 39672391
Publications in 2023
Bhutada S, Tran-Lundmark K, Kramer B, Conner P, Lowry AM, Blackstone E, Frenckner B, Mesas-Burgos C, Apte SS. Identification of protein biomarkers associated with congenital diaphragmatic hernia in human amniotic fluid. Sci Rep. 2023 Sep 19;13(1):15483. doi: 10.1038/s41598-023-42576-2. PMID: 37726509; PMCID: PMC10509251.
Nandadasa S, Martin D, Deshpande G, Robert KL, Stack MS, Itoh Y, Apte SS. Degradomic Identification of Membrane Type 1-Matrix Metalloproteinase as an ADAMTS9 and ADAMTS20 Substrate. Mol Cell Proteomics. 2023 Jun;22(6):100566. doi: 10.1016/j.mcpro.2023.100566. Epub 2023 May 9. PMID: 37169079; PMCID: PMC10267602.
Bhutada S, Apte SS: The Proteolytic Landscape Of Healthy Juvenile Bovine Knee Cartilage And Synovial Fluid As A Comparative Resource For The Human Osteoarthritis Degradome. Osteoarthritis and Cartilage. 2023 March(2). doi: 10.1016/j.joca.2023.01.396
McConaghy K, Klika AK, Apte SS, Erdemir A, Derwin K, Piuzzi NS. A Call to Action for Musculoskeletal Research Funding: The Growing Economic and Disease Burden of Musculoskeletal Conditions in the United States Is Not Reflected in Musculoskeletal Research Funding. J Bone Joint Surg Am. 2023 Mar 15;105(6):492-498. doi: 10.2106/JBJS.22.00693. Epub 2022 Nov 9. PMID: 36574617.
van der Have, O, Mead, TJ, Westöö, C, Peruzzi, N, Mutgan, AC, Norvik, C, Bech, M, Struglics, A, Hoetzenecker, K, Brunnström, H, Westergren-Thorsson, G, Kwapiszewska, G, Apte, SS, Tran-Lundmark, K. Aggrecan accumulates at sites of increased pulmonary arterial pressure in idiopathic pulmonary arterial hypertension. Pulm Circ. 2023; 13:e12200. https://doi.org/10.1002/pul2.12200
Apte, S.S., 2023. ADAMTS proteases: mediators of physiological and pathogenic extracellular proteolysis. 808-819. Elsevier. https://doi.org/10.1016/B978-0-12-394447-4.10074-4
Publications in 2022
Apte SS, Naba A. Beyond the Matrisome: New Frontiers in ECM Research. Matrix Biol. 2022 Dec 23:S0945-053X(22)00152-4. doi: 10.1016/j.matbio.2022.12.003. Epub ahead of print. PMID: 36572230.
Rypdal KB, Olav Melleby A, Robinson EL, Li J, Palmero S, Seifert DE, Martin D, Clark C, López B, Andreassen K, Dahl CP, Sjaastad I, Tønnessen T, Stokke MK, Louch WE, González A, Heymans S, Christensen G, Apte SS, Lunde IG. ADAMTSL3 knock-out mice develop cardiac dysfunction and dilatation with increased TGFβ signaling after pressure overload. Commun Biol. 2022 Dec 20;5(1):1392. doi: 10.1038/s42003-022-04361-1. PMID: 36539599; PMCID: PMC9767913.
Foulcer SJ, Day AJ, Apte SS. Isolation and Purification of Versican and Analysis of Versican Proteolysis. Methods Mol Biol. 2022;2303:559-578. doi: 10.1007/978-1-0716-1398-6_43. PMID: 34626407.
Kuwabara JT, Hara A, Bhutada S, Gojanovich GS, Chen J, Hokutan K, Shettigar V, Lee AY, DeAngelo LP, Heckl JR, Jahansooz JR, Tacdol DK, Ziolo MT, Apte SS, Tallquist MD. Consequences of PDGFRα+ fibroblast reduction in adult murine hearts. Elife. 2022 Sep 23;11:e69854. doi: 10.7554/eLife.69854. PMID: 36149056; PMCID: PMC9576271.
Kuwabara JT, Hara A, Heckl JR, Peña B, Bhutada S, DeMaris R, Ivey MJ, DeAngelo LP, Liu X, Park J, Jahansooz JR, Mestroni L, McKinsey TA, Apte SS, Tallquist MD. Regulation of extracellular matrix composition by fibroblasts during perinatal cardiac maturation. J Mol Cell Cardiol. 2022 Aug;169:84-95. doi: 10.1016/j.yjmcc.2022.05.003. Epub 2022 May 13. PMID: 35569524.
Bhutada S, Li L, Willard B, Muschler G, Piuzzi N, Apte SS. Forward and reverse degradomics defines the proteolytic landscape of human knee osteoarthritic cartilage and the role of the serine protease HtrA1. Osteoarthritis Cartilage. 2022 Aug;30(8):1091-1102. doi: 10.1016/j.joca.2022.02.622. Epub 2022 Mar 24. PMID: 35339693.
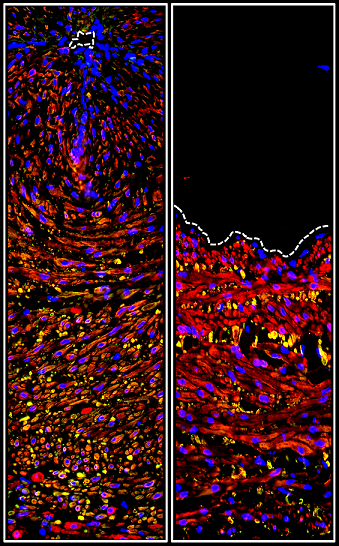
Perry WJ, Grunenwald CM, Van de Plas R, Witten JC, Martin DR, Apte SS, Cassat JE, Pettersson GB, Caprioli RM, Skaar EP, Spraggins JM. Visualizing Staphylococcus aureus pathogenic membrane modification within the host infection environment by multimodal imaging mass spectrometry. Cell Chem Biol. 2022 Jul 21;29(7):1209-1217.e4. doi: 10.1016/j.chembiol.2022.05.004. Epub 2022 Jun 1. PMID: 35654040; PMCID: PMC9308753.
Mead TJ, Bhutada S, Martin DR, Apte SS. Proteolysis: a key post-translational modification regulating proteoglycans. Am J Physiol Cell Physiol. 2022 Sep 1;323(3):C651-C665. doi: 10.1152/ajpcell.00215.2022. Epub 2022 Jul 4. PMID: 35785985; PMCID: PMC9448339.
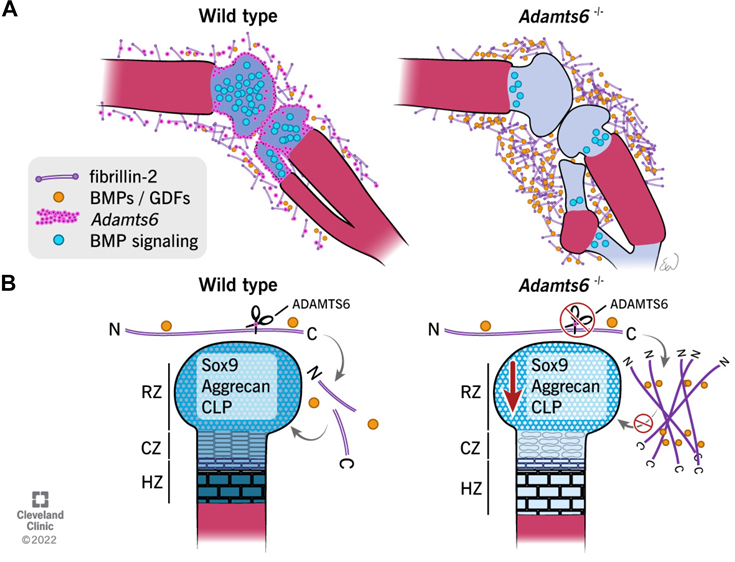
Mead TJ, Martin DR, Wang LW, Cain SA, Gulec C, Cahill E, Mauch J, Reinhardt D, Lo C, Baldock C, Apte SS. Proteolysis of fibrillin-2 microfibrils is essential for normal skeletal development. Elife. 2022 May 3;11:e71142. doi: 10.7554/eLife.71142. PMID: 35503090; PMCID: PMC9064305.
Witten JC, Tan CD, Rodriguez ER, Shrestha NK, Gordon SM, Hussain ST, Apte SS, Unai S, Blackstone EH, Pettersson GB. Invasive Aortic Valve Endocarditis: Clinical and Tissue Findings From a Prospective Investigation. Ann Thorac Surg. 2022 Feb;113(2):535-543. doi: 10.1016/j.athoracsur.2021.03.072. Epub 2021 Apr 8. PMID: 33839129.
Publications in 2021
Rypdal KB, Erusappan PM, Melleby AO, Seifert DE, Palmero S, Strand ME, Tønnessen T, Dahl CP, Almaas V, Hubmacher D, Apte SS, Christensen G, Lunde IG. The extracellular matrix glycoprotein ADAMTSL2 is increased in heart failure and inhibits TGFβ signaling in cardiac fibroblasts. Sci Rep. 2021 Oct 5;11(1):19757. doi: 10.1038/s41598-021-99032-2.
Martin DR, Santamaria S, Koch CD, Ahnström J, Apte SS. Identification of novel ADAMTS1, ADAMTS4 and ADAMTS5 cleavage sites in versican using a label-free quantitative proteomics approach. J Proteomics. 2021 Aug 24:104358. doi: 10.1016/j.jprot.2021.104358. Online ahead of print.
Apte SS. The Pivotal Role of Versican Turnover by ADAMTS Proteases in Mammalian Reproduction and Development. In Proteoglycans in Stem Cells. From Development to Cancer. Götte, M, Forsberg-Nilsson, K (Eds.), Springer 2021, ISBN 978-3-030-73452-7.
Nandadasa S, Burin des Rozier C, Koch C, Tran-Lundmark K, Dours-Zimmermann MT, Zimmermann DR, Valleix S, Apte SS. A new mouse mutant with cleavage-resistant versican and isoform-specific versican mutants demonstrate that proteolysis at the Glu441-Ala442 peptide bond in the V1 isoform is essential for interdigital web regression. Matrix Biology Plus, 2021, May 14;10:100064 | doi: 10.1016/j.mbplus.2021.100064.
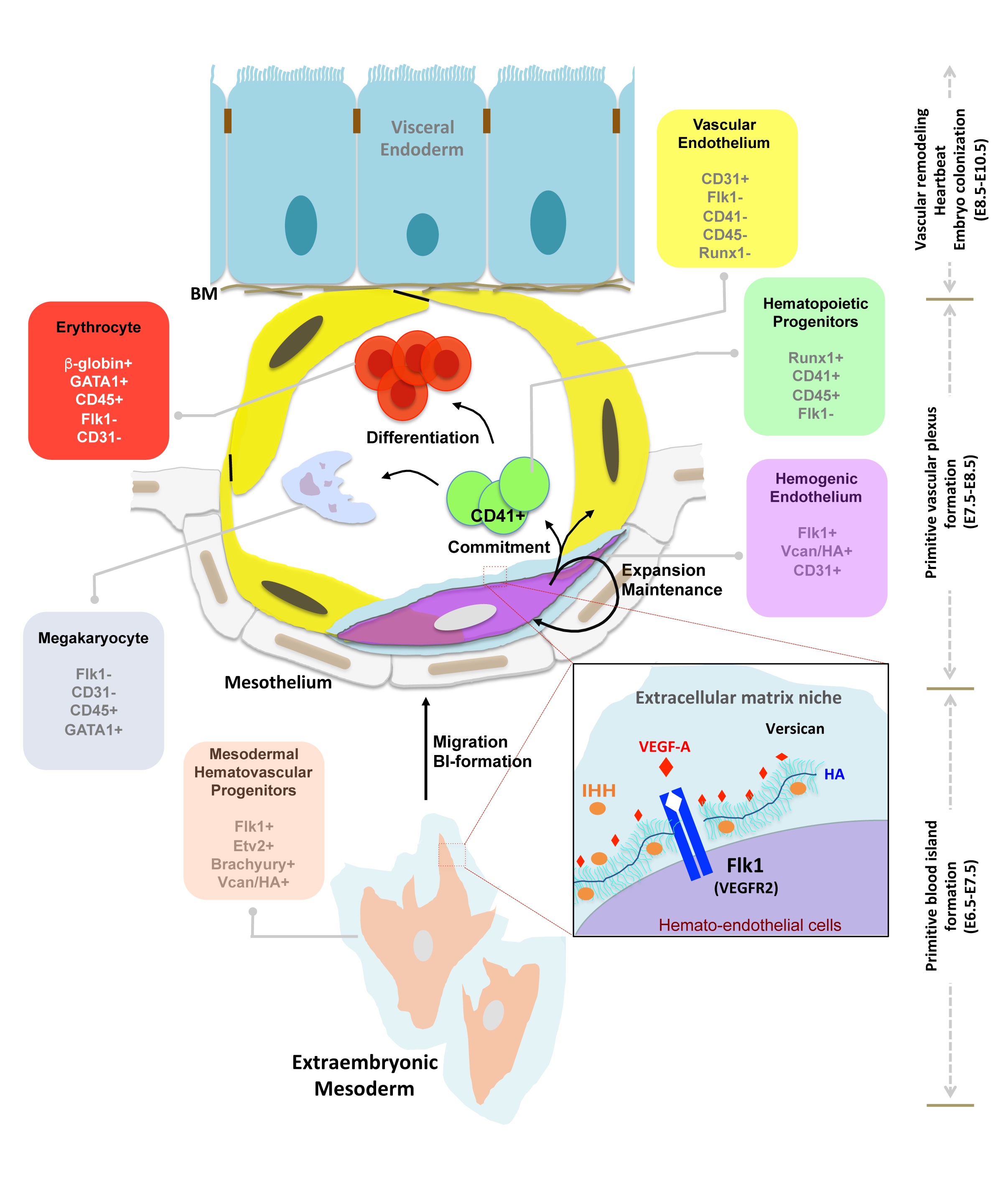
Nandadasa S, O'Donnell A, Murao A, Yamaguchi Y, Midura RJ, Olson L, Apte SS. The versican-hyaluronan complex provides an essential extracellular matrix niche for Flk1+ hematoendothelial progenitors. Matrix Biol. 2021 Mar;97:40-57 | doi: 10.1016/j.matbio.2021.01.002.
Arechederra M, Bazai SK, Abdouni A, Sequera C, Mead TJ, Richelme S, Daian F, Audebert S, Dono R, Lozano A, Gregoire D, Hibner U, Allende DS, Apte SS, Maina F. ADAMTSL5 is an epigenetically activated gene underlying tumorigenesis and drug resistance in hepatocellular carcinoma. J Hepatol. 2021 Apr;74(4):893-906 | doi: 10.1016/j.jhep.2020.11.008.
Foulcer SJ, Day AJ, Apte SS. Isolation and Purification of Versican and Analysis of Versican Proteolysis. Methods Mol Biol. 2022;2303:559-578 | doi: 10.1007/978-1-0716-1398-6_43
Balic Z, Misra S, Willard B, Reinhardt DP, Apte SS, Hubmacher D. Alternative splicing of the metalloprotease ADAMTS17 spacer regulates secretion and modulates autoproteolytic activity. FASEB J. 2021 Feb;35(2):e21310 doi: 10.1096/fj.202001120RR.
Publications in 2020
Nandadasa S, Szafron JM, Pathak V, Murtada S-I, Kraft CM, O ’ Donnell A, Norvik C, Hughes C, Caterson B, Domowicz MS, Schwartz NB, Tran-Lundmark K, Veigl M, Sedwick D, Philipson EH, Humphrey JD, Apte SS. Vascular dimorphism ensured by regulated proteoglycan dynamics favors rapid umbilical artery closure at birth. Accompanying Commentary: https://elifesciences.org/articles/63128. Elife. 2020 Sep 10;9:e60683 | doi: 10.7554/eLife.60683.
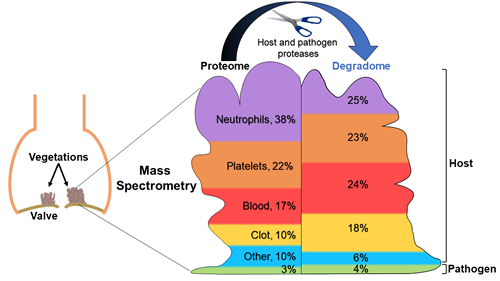
Martin DR, Witten JC, Tan CD, Rodrigues ER, Pettersson GB, Seifert DE, Willard BB, Apte SS. Proteomics identifies a convergent innate response to infective endocarditis and extensive proteolysis in vegetation components. JCI Insight 2020 Jun 16;135317 | doi: 10.1172/jci.insight.135317Ataca D, Aouad P, Constantin C, Laszlo C, Beleut M, Shamseddin M, Rajaram RD, Jeitziner R, Mead TJ, Caikovski M, Bucher P, Ambrosini G, Apte SS, Brisken C. The secreted protease ADAMTS18 links hormone action to activation of the mammary stem cell niche. Nat Commun. 2020 Mar 26;11(1):1571 | doi: 10.1038/s41467-020-15357-y.
Zhang A, Berardinelli SJ, Leonhard-Melief C, Vasudevan D, Liu TW, Taibi A, Giannone S, Apte SS, Holdener BC, Haltiwanger RS. O-Fucosylation of ADAMTSL2 is required for secretion and is impacted by geleophysic dysplasia-causing mutations. J Biol Chem. 2020 Sep 10:jbc.RA120.014557 | doi: 10.1074/jbc.RA120.014557.
Evans DR, Green JS, Fahiminiya S, Majewski J, Fernandez BA, Deardorff MA, Johnson GJ, Whelan JH, Hubmacher D, Apte SS; Care4Rare Canada Consortium, Woods MO. A novel pathogenic missense ADAMTS17 variant that impairs secretion causes Weill-Marchesani Syndrome with variably dysmorphic hand features. Sci Rep. 2020 Jul 2;10(1):10827. | doi: 10.1038/s41598-020-66978-8
Koch CD, Lee CM, Apte SS. Aggrecan in Cardiovascular Development and Disease. J Histochem Cytochem. 2020 Sep 1:22155420952902. | doi: 10.1369/0022155420952902.
Koch CD, Apte SS. Characterization of Proteoglycanomes by Mass Spectrometry. In, S. Ricard-Blum (ed.) Extracellular Matrix Omics, In the series, Biology of Extracellular Matrix, pp69-82. ISBN 978-3-030-58329-3 7. SpringerNature Switzerland AG 2020 | https://doi.org/10.1007/978-3-030-58330-9_4
Apte SS. ADAMTS Proteins: Concepts, Challenges, and Prospects. Methods Mol Biol. 2020;2043:1-12 | doi: 10.1007/978-1-4939-9698-8_1. Review
Mead TJ, Apte SS. Visualization and Quantification of Pericellular Matrix. Methods Mol Biol. 2020;2043:261-264 | doi: 10.1007/978-1-4939-9698-8_21
Mead TJ, Apte SS. Expression Analysis by RNAscope™ In Situ Hybridization. Methods Mol Biol. 2020;2043:173-178 | doi: 10.1007/978-1-4939-9698-8_14
Publications in 2019
Wang LW, Nandadasa S, Annis DS, Dubail J, Mosher DF, Willard BB, Apte SS. A disintegrin-like and metalloproteinase domain with thrombospondin type 1 motif 9 (ADAMTS9) regulates fibronectin fibrillogenesis and turnover. J Biol Chem, 2019,294:9924-9936 | doi: 10.1074/jbc.RA118.006479
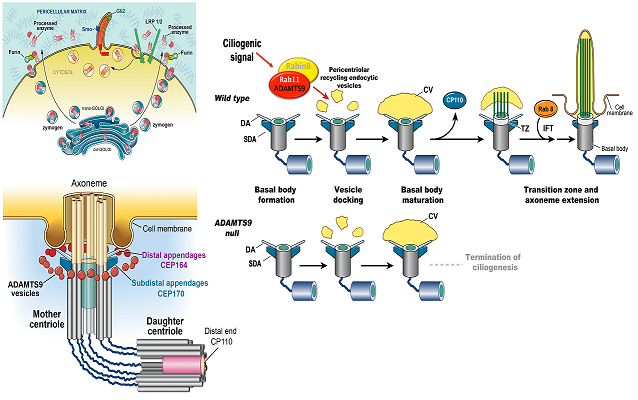
Hubmacher, D, Thacker, S, Adams SM, Birk, D, Schweitzer, R, Apte, SS. Limb- and tendon-specific Adamtsl2 deletion identifies a soft tissue mechanism modulating bone length. Matrix Biology, 2019. pii: S0945-053X(18)30372-X | doi: 10.1016/j.matbio.2019.02.001.Nandadasa, S, Kraft, CM, O’Donnell, A, Wang, LW, O’Donnell, A, Patel, R, Gee HY, Grobe, K, Cox, TC, Hildebrandt F and Apte, SS. Secreted metalloproteases ADAMTS9 and ADAMTS20 have a non-canonical role in ciliary vesicle growth during ciliogenesis. Nature Communications, (2019) 10:953 | https://doi.org/10.1038/s41467-019-08520-7
Choi YJ, Halbritter J, Braun DA, Schueler M, Schapiro D, Rim JH, Nandadasa S, Choi WI, Widmeier E, Shril S, Körber F, Sethi SK, Lifton RP, Beck BB, Apte SS, Gee HY,Hildebrandt F. Mutations of ADAMTS9 Cause Nephronophthisis-Related Ciliopathy. Am J Hum Genet. 2019 104:45-54. | doi: 10.1016/j.ajhg.2018.11.003.
Wang LW, Kutz WE, Mead TJ, Beene LC, Singh S, Jenkins MW, Reinhardt DP, Apte SS. Adamts10 inactivation in mice leads to persistence of ocular microfibrils subsequent to reduced fibrillin-2 cleavage. Matrix Biol. 2019, 77:117-128. pii: S0945-053X(18)30253-1. doi: 10.1016/j.matbio.2018.09.004.
Jensen LD, Hot B, Ramsköld D, Germano RFV, Yokota C, Giatrellis S, Lauschke VM, Hubmacher D, Li MX, Hupe M, Arnold TD, Sandberg R, Frisén J, Trusohamn M, Martowicz A, Wisniewska-Kruk J, Nyqvist D, Adams RH, Apte SS, Vanhollebeke B, Stenman JM, Kele J. Disruption of the Extracellular Matrix Progressively Impairs Central Nervous System Vascular Maturation Downstream of β-Catenin Signaling. Arterioscler Thromb Vasc Biol. 2019 39:1432-1447. doi: 10.1161/ATVBAHA.119.312388.
Santamaria S, Yamamoto K, Teraz-Orosz A, Koch C, Apte SS, de Groot R, Lane DA, Ahnström J. Exosites in Hypervariable Loops of ADAMTS Spacer Domains control Substrate Recognition and Proteolysis. Sci Rep. 2019 Jul 29;9(1):10914. doi: 10.1038/s41598-019-47494-w
Holdener BC, Percival CJ, Grady RC, Cameron DC, Berardinelli SJ, Zhang A, Neupane S, Takeuchi M, Jimenez-Vega JC, Uddin SMZ, Komatsu DE, Honkanen R, Dubail J, Apte SS, Sato T, Narimatsu H, McClain SA, Haltiwanger RS. ADAMTS9 and ADAMTS20 are differentially affected by loss of B3GLCT in a mouse model of Peters Plus Syndrome. Hum Mol Genet. 2019 Oct 10. pii: ddz225. doi: 10.1093/hmg/ddz225. [Epub ahead of print]
Graae AS, Grarup N, Ribel-Madsen R, Lystbæk SH, Boesgaard T, Staiger H, Fritsche A, Wellner N, Sulek K, Kjolby M, Backe MB, Chubanava S, Prats C, Serup AK, Birk JB, Dubail J, Gillberg L, Vienberg SG, Nykjær A, Kiens B, Wojtaszewski JFP, Larsen S, Apte SS, Häring HU, Vaag A, Zethelius B, Pedersen O, Treebak JT, Hansen T, Holst B.ADAMTS9 Regulates Skeletal Muscle Insulin Sensitivity Through Extracellular Matrix Alterations. Diabetes. 68(3):502-514. doi: 10.2337/db18-0418.
A sampling of research publications from 2018 and earlier
1. Mead TJ, Du Y, Nelson CM, Gueye N-A, Drazba J, Dancevic CM, Vankemmelbeke M, Buttle DJ, Apte SS. ADAMTS9-Regulated Pericellular Matrix Dynamics Governs Focal Adhesion-Dependent Smooth Muscle Differentiation. Cell Reports. 2018, 23; 2, 485-498.
2. Cikach FS, Koch CD, Mead TJ, Galatioto J, Willard BB, Emerton KB, Eagleton MJ, Blackstone EH, Ramirez F, Roselli EE, Apte SS. Massive aggrecan and versican accumulation in thoracic aortic aneurysm and dissection. JCI Insight. 2018 Mar 8;3(5). pii: 97167. doi: 10.1172/jci.insight.97167.
3. Prins, B. , Mead, T.J. (joint first authors) et al. Exome-chip meta-analysis identifies novel loci associated with cardiac conduction, including ADAMTS6. Genome Biol 2018, 19:87. https://doi.org/10.1186/s13059-018-1457-6
4. Hubmacher D, Schneider M, Berardinelli S, Takeuchi H, Willard B, Reinhardt DH, Haltiwanger R, and Apte SS. Unusual life cycle and impact on microfibril assembly of ADAMTS17, a secreted metalloprotease mutated in genetic eye disease. Scientific Reports, 2017;7:41871. doi: 10.1038/srep41871.
5. Dubail, J, Vasudevan, D, Wang, LW, Earp, SE, Jenkins, MW, Haltiwanger, RS, and Apte SS. Impaired ADAMTS9 secretion: A potential mechanism for eye defects in Peters Plus Syndrome. Scientific Reports. 2016. 6:33974. doi: 10.1038/srep33974.
6. Nandadasa, S., Nelson, C.M., Apte SS. ADAMTS9-Mediated Extracellular Matrix Dynamics Regulates Umbilical Cord Vascular Smooth Muscle Differentiation and Rotation. Cell Reports 11:1519-28, 2015
7. Enomoto, H., Nelson, C., Somerville, R.P.T., Mielke, K., Dixon, L., Powell, K., Apte SS. Cooperation of two ADAMTS metalloproteases in closure of the mouse palate identifies a requirement for versican proteolysis in regulating palatal mesenchyme proliferation. Development, 2010, 37:4029-38.
8. McCulloch, D.R., Nelson, C.M., Dixon, L.J., Silver D.L., Wylie, J.D., Lindner, V., Sasaki, T., Cooley, M.A., Argraves, W.S. and Apte SS. ADAMTS metalloproteases generate active versican fragments that regulate interdigital web regression. Developmental Cell 2009, 17:687-98. PMCID:PMC2780442.
Books
Apte SS. ADAMTS proteins: Concepts, Challenges and Prospects. Methods Mol Biol. 2020;2043:1-12. doi: 10.1007/978-1-4939-9698-8_1.
Reviews, technical reports and commentaries
1. Mead TJ, Apte SS. ADAMTS proteins in human disorders.Matrix Biol. 2018 Jun 6. pii: S0945-053X(18)30179-3. doi: 10.1016/j.matbio.2018.06.002.
2. Apte, SS. Anti-ADAMTS5 monoclonal antibodies: implications for aggrecanase inhibition in osteoarthritis. Biochem J. 2016 Jan 1;473(1):e1-e4
3. Dubail J, Apte SS. Insights on ADAMTS proteases and ADAMTS-like proteins from mammalian genetics.Matrix Biol. 44-46:24-37 2015
4. Foulcer SJ, Day AJ, Apte SS. Isolation and purification of versican and analysis of versican proteolysis. Methods Mol Biol. 1229:587-604, 2015
5. Nandadasa S, Foulcer S, Apte SS. The multiple, complex roles of versican and its proteolytic turnover by ADAMTS proteases during embryogenesis. Matrix Biol. 2014, 35:34-41, 2014.
Book chapters
1. Apte, SS. Chapter 259. Connective Tissue Structure and Function. ed. Goldman L, and Shafer, A.I., Goldman-Cecil Textbook of Medicine, Twenty-Fifth Edition, Elsevier, New York, 2016, ISBN 9781455750177
2. Apte SS. ADAMTS proteases: Mediators of physiological and pathogenic extracellular proteolysis, in Bradshaw, R., and Stahl, P, eds, Encyclopedia of Cell Biology, Elsevier, New York, 2015, ISBN 9780123944474
3. Apte, SS. Chapter 2. Overview of the ADAMTS superfamily, in Rodgers, G., ed, ADAMTS13: Biology and Disease, Springer, New York, 2015, ISBN 9783319087160
4. Traboulsi I, and Apte SS. Chapter 43. Ectopia Lentis and Associated Systemic Disease, in Traboulsi, I., ed, Genetic Diseases of the Eye, Second Edition, Oxford University Press, USA, 2012, ISBN 9780195326147
Patents
| US Patent | Patent Title | Issue Date | First-Named Inventor |
|---|---|---|---|
| 6,391,610 | Nucleic Acids Encoding Zinc Metalloproteases | 05/21/2002 | Suneel S. Apte, PhD |
Research News

The design team behind DICED answer questions about the first-of-its-kind searchable web interface for specialized proteomics data.

A multidisciplinary team of researchers, led by Dr. Suneel Apte, identified a new biomarker for osteoarthritis that may contribute to cartilage damage and destruction.

Researchers have discovered that umbilical cord arteries contain components that naturally promote narrowing at birth, and ensure a sequence in which the arteries close before the vein, supporting delayed cord clamping following delivery in most uncomplicated deliveries.

A team of researchers, led by the Department of Biomedical Engineering, found that differences in the protein makeup of vegetations may help to identify the pathogen of infective endocarditis and cast new light upon the deadly disease.

Researchers used genetic engineering and gene editing technology to inactivate genes that produce the proteases ADAMTS9 and ADAMTS20, leading to birth defects including faulty closure of the neural tube and palate and failed separation of the trachea and esophagus.

Researchers used specialized assays to detect five new gene variants with particularly low frequencies (minor allele frequency of less than 1%) in coding regions of the DNA.

The multidisciplinary team of researchers silenced ADAMTS9 in a preclinical model and cultured human uterine SMCs.